RStudio
RStudio is a powerful interactive environment dedicated to the R programming language. To utilize RStudio within Open OnDemand, follow these steps:
Make sure you don’t have any commands that produce output in your $HOME/.bashrc file. Run this command:
source $HOME/.bashrcand observe if output is produced. If so, remove the command that produces output by editing the file $HOME/.bashrc.
- Select Interactive Apps from the main menu and then click on "R Studio Server".

- Fill out the required session configuration form:
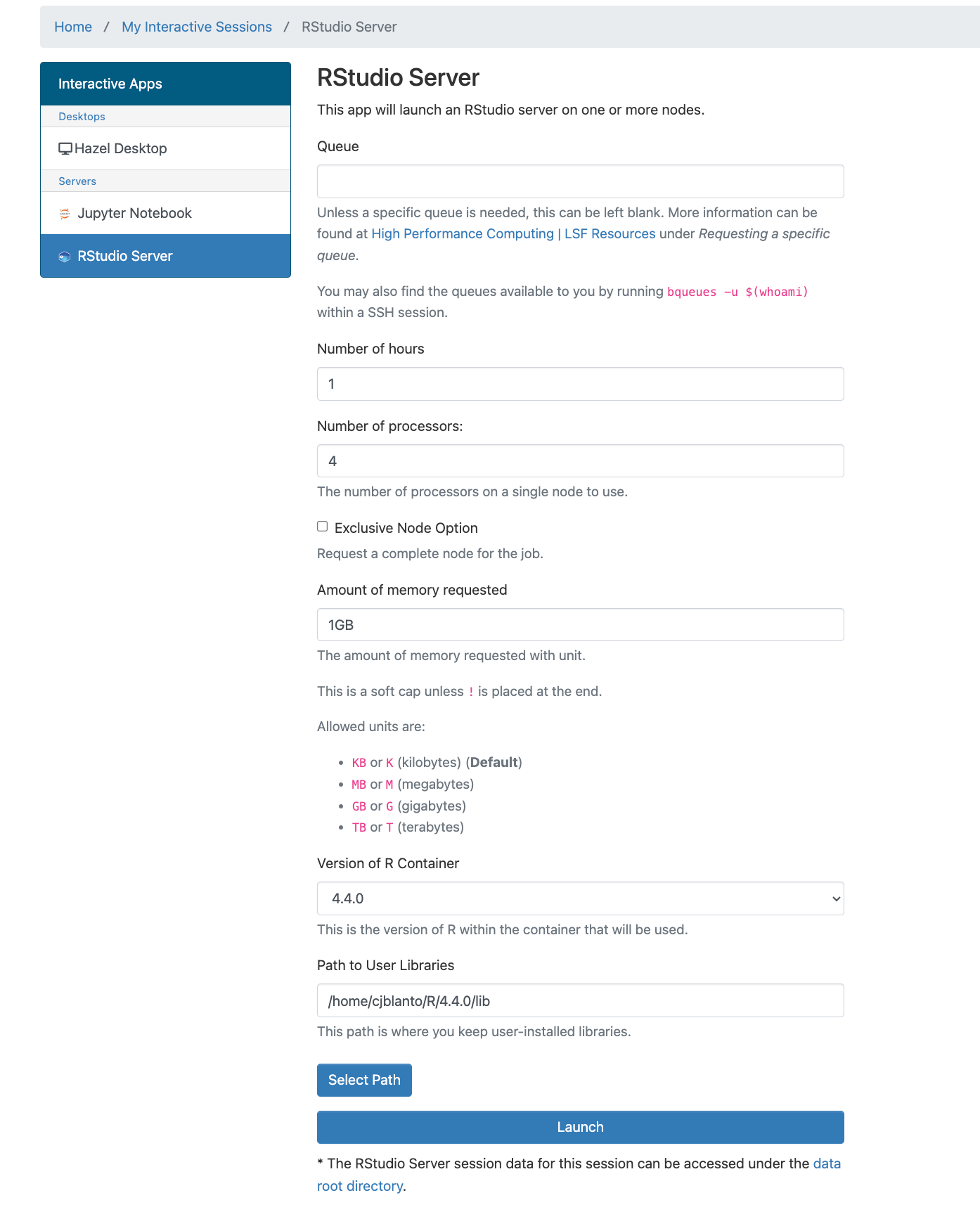 The various parameters mean:
The various parameters mean:
- Queue: Usually, this field can be left empty unless a specific queue needs to be selected.
- Number of hours: Indicates the duration for which the session will be active.
- Number of processors: Specifies how many processors the session will utilize.
- Exclusive Node Option: Requests exclusive access to a node; select multiple processors for scheduling, understanding that it may delay job initiation.
- Amount of memory requested: Specify memory requirements, such as “1GB” for one gigabyte.
- Version of R container: Select the appropriate R version for the session.
- Path to User Libraries: Define where user-installed libraries are stored; it's advised to separate this location from standard libraries to prevent conflicts.
- After finalizing the configuration, click on the Launch button. Job initiation time may vary based on resource allocation. Once ready, select the Connect to RStudio Server button to access the RStudio application.
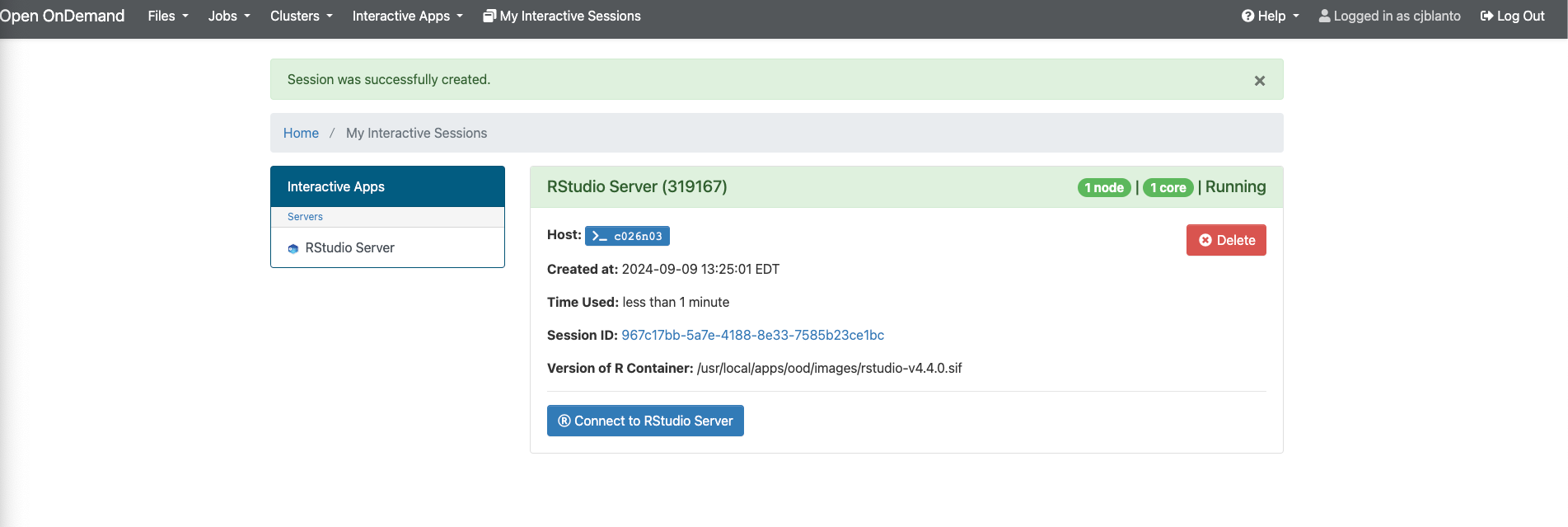
Working within RStudio
Upon clicking the Connect to RStudio Server button, the RStudio application interface will appear, offering familiar functionalities and features.
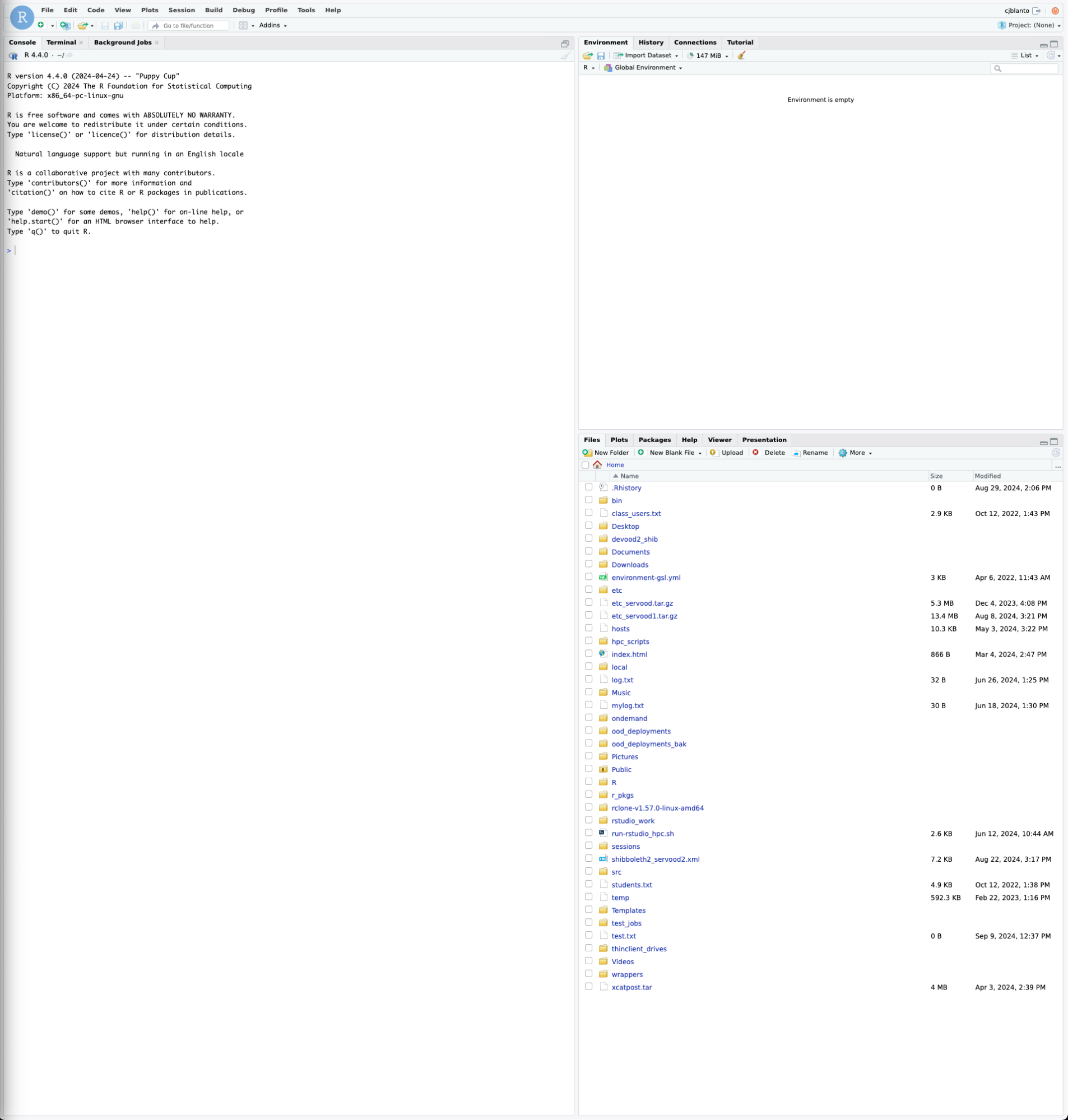 Users are encouraged to utilize RStudio as they typically would in local environments with a few caveats.
Users are encouraged to utilize RStudio as they typically would in local environments with a few caveats.
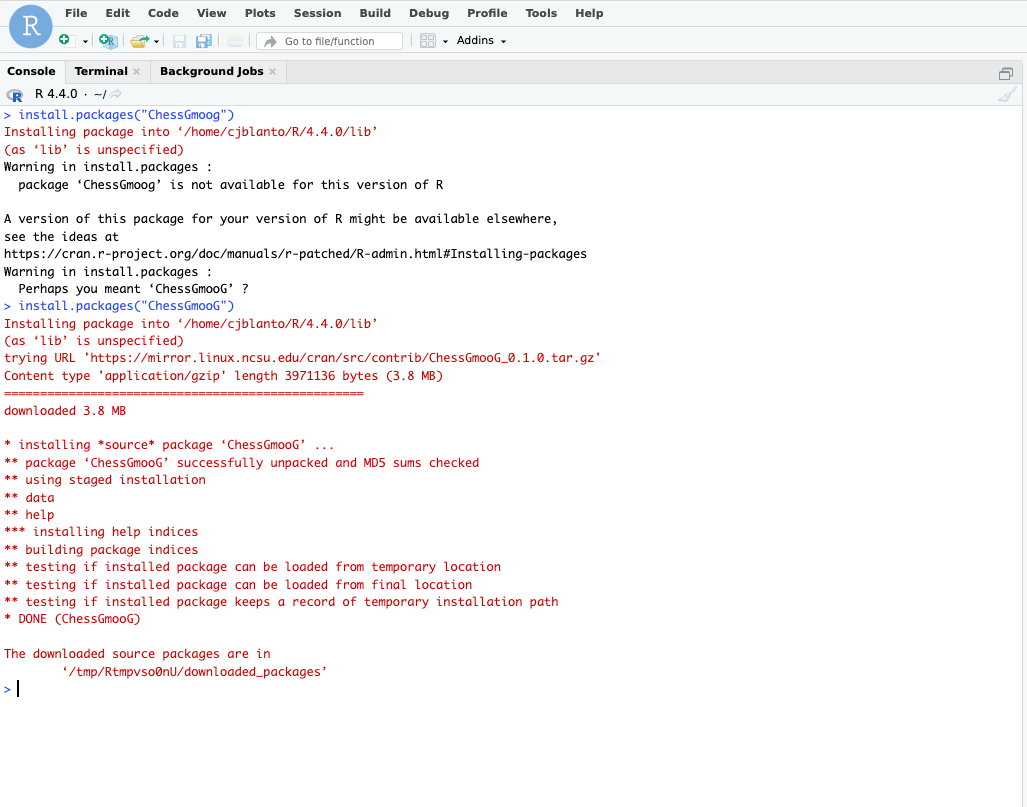
Installing Packages
The version of RStudio utilized in the compute nodes supports package installation. Packages can be sourced from the NC State CRAN mirror or the Duke CRAN mirror.
Resuming Interrupted Sessions
In instances where your session is still active, you can re-establish the connection (useful in case of broken connections) via the My Interactive Sessions menu.
 Any currently running sessions will be listed, and users can reconnect by clicking the Connect to RStudio Server button.
Any currently running sessions will be listed, and users can reconnect by clicking the Connect to RStudio Server button.
Exiting RStudio
To properly exit your RStudio session, follow these steps:
- Click the Power button located in the RStudio interface.

- Close the RStudio window to end your session.
- Finally, back in the My Interactive Session window, click the Delete option to remove the session from active job listings.
Limitations of OOD RStudio
While Open OnDemand with RStudio offers substantial advantages, it does come with inherent limitations, including:
- Limited Internet Connectivity: Not all websites and services may be accessed, impacting functionalities that rely on external resources.
- Restricted CRAN Mirrors: Access to CRAN mirrors is limited, which may affect package installations.
- Potential Configuration Conflicts: Some of the user configurations from local environments may clash with the OOD version settings, leading to unexpected behaviors.
- Missing Full Internet Connection Resources: Certain components, particularly those requiring a full internet connection such as the Connections panel (ODBC, Spark), may encounter errors or be non-operational.
- Inability to Install Certain R Packages: Packages that have compiled components, such as shiny or learnr, cannot be installed due to absent dependencies and components required for configuration.
Troubleshooting
502 Errors
If you encounter a 502 Proxy error, it is advisable to check the connection speed of your account logins via the SSH Shell. Slow login times can often lead to Open OnDemand timeout issues. A common culprit is the inclusion of certain commands within the .bashrc file, which may significantly delay login speeds.
For example, users who have added code for Anaconda will need to remove the code from the .bashrc file and relocate it to an alternative file, such as .conda.bashrc, for efficient sourcing when necessary.
Last modified: April 27 2025 01:42:06.